
Blog: Sequencing Technologies in the Food Safety Industry
Next-generation sequencing (NGS) has revolutionized the speed and accuracy of genetic testing and identification. While primarily associated with healthcare, NGS is beginning to play an important role in food safety—the advanced technology can quickly identify pathogens with previously unparalleled accuracy. While the integration of NGS into present-day food safety systems is yet to be complete, government agencies have found frequent and valuable use in the technology for food-borne disease diagnostics, safety inspections, and outbreak tracing.
The Science of Food Safety
The transmission of pathogens through food has considerable costs to human health and society—the annual economic burden of these pathogens is estimated at $17.6 billion (in 2018 dollars) according to the U.S. Department of Agriculture (USDA). Food-borne pathogens are also particularly notorious for their ability to spread widely at a rapid pace. With improved supply chain management facilitating expanded reach of individual food supplies, a single source can affect a number of different locations, making quick identification and accurate tracking of pathogens increasingly imperative for disease prevention.
The adoption of whole-genome sequencing (WGS) using high throughput NGS has expanded our capacity to sequence the entire bacterial genome quickly and cheaply, allowing us to obtain previously unheard-of precision information about a pathogen at the DNA and gene levels.
Although most of the current uses in food safety for WGS data is retrospectively in pathogen source detection, there are several ways it can be used in a predictive manner as well. Beyond predicting shelf life and spoilage of food, WGS data can also be used to predict resistance to antimicrobials or biocides.

Barriers to Adoption
Despite its potential, high-throughput NGS has not yet been fully incorporated into existing food safety systems. While the application of this new technology for illness diagnosis, food safety checks, and epidemic investigations is widespread, the food industry’s adoption of this sequencing technique has been comparatively slow and fragmented.
Even though WGS can conclusively confirm or deny possible root causes, there are still significant barriers to adoption including cost, complexity, and the time it takes to get results. Although the cost of sequencing has dropped substantially it is still more expensive than other subtyping tools due to investments required in IT infrastructure, laboratory equipment and the development of expertise in sequencing, bioinformatics and microbial genomics.

Additional challenges for the adoption of WGS in food safety are the complexity and time it takes to complete the analysis, a major drawback of WGS is that it is performed on cultured organisms and that sequencing at scale can take tens of hours to days. The resulting data requires interpretation that must account for variation in lab preparation, a wide variety of bioinformatic tools and parameterization of said tools, and the unknown complexities of microbial evolution.
A final barrier to uptake in the food industry is the legal uncertainty caused by the absence of a clear legal and regulatory framework for sharing WGS data and the methodology used by global regulatory agencies in evaluating it. The industry has expressed concern regarding misinterpretation of the data and the potential legal risks and litigation impacts. A potential step toward a solution is the proposal for a common database of laboratory positive control and PTS strains for the benefit of all companies.
Future of NGS in Food Safety
Despite the barriers, the number of global food manufacturers investing in high throughput sequencing (HTS) technology has grown steadily in the past few years and is being used for tasks such as source tracking, environmental surveys and monitoring, as well as R&D evaluations into genetic responses to heat stress and characterization of foodborne pathogenic bacteria in the food supply chain.
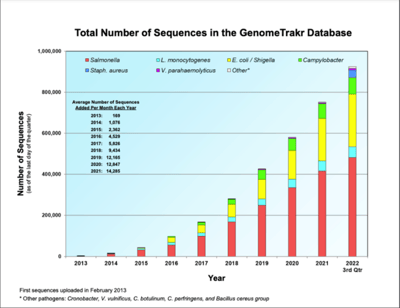
While full sequencing and analysis infrastructure has only been adopted by a few large food companies thus far, there are cost efficient ways for lower-capacity food producers to also utilize this technology. Data can be analyzed using small sequencers and a fast internet connection in conjunction with commercially available bioinformatics applications like Almaden’s g.nome® platform. Extensive WGS datasets are housed and available to the public in the FDA’s Center for Food Safety and Applied Nutrition’s GenomeTrakr database for foodborne and environmental bacterial pathogens. This distributed network of federal labs, state health and university labs, hospital labs and other labs outside the United States have sequenced more than 924,000 isolates and closed more than 300 genomes.
Diagnostics are increasingly being done using molecular methods without culture, yielding fewer isolates for sequencing in the future. This trend is progressing work in metagemomics where isolate-level data is obtained without culturing by sequencing all DNA in a specimen. This area of work is showing promise for not only replacing isolate-based sequencing, but also for documenting the authenticity of food and the presence of contaminants by gathering information about the microbial populations in feed, animals, food, people and the environment.
Advancements in NGS technology have had a significant impact on the life sciences industry, bringing costs down and paving the path from analytical microbiology in the research lab to a genetic testing kit delivered to the doorstep of consumers. It is interesting to think about how that journey might be mirrored in the food industry. WGS is expected to remain a fixture in public health food safety management, but adoption by the broader food industry is still complicated and currently only being used when alternate subtyping methods are not sufficient to differentiate isolates. Food companies would be wise to keep pace with regulatory agencies as their interest and use of WGS continues to intensify, or they risk being behind the regulatory curve.
Food safety is undoubtably being modernized by next generation sequencing and analysis. Although there remain significant barriers to widespread adoption, the promise of a safer global food supply chain is an endeavor that we can all support.
For the analysis of sequencing data to keep up with the scale and application of these new technologies, modern companies need to be able to generate workflows that are scalable, flexible enough to be re-purposed for new and related problems, reproducible to derive confident results, auditable for oversight, and portable so that they can become standardized.
Learn more about how g.nome can help streamline your genomic workflows. Brought to market by a team of bioinformaticians, engineers and industry professionals, g.nome provides a low-code/no-code pipeline build. With a visual drag-and-drop pipeline builder that utilizes toolkits and pre-built workflows from a curated library, g.nome is giving power to researchers — enabling them to import custom code, handle big datasets reliably and optimize team collaboration from anywhere.
With g.nome, long-time barriers linked to workflow language, process flow visibility and quality control are removed. All that’s left are streamlined, scalable and interoperable genomic workflows — leaving research teams to do what they do best: focus on the science.
Mark Kunitomi
Mark is the Chief Scientific Officer for Almaden Genomics. He was a post-doctoral fellow at UC San Francisco with a background in genomics, bioinformatics and microbiology. He has a Ph.D. in Biochemistry & Molecular Biology from the University of California, San Francisco. Click to learn more about Almaden Genomics and the g.nome platform.