SINGLE-CELL RNA-SEQ
Revolutionize scRNA-seq analysis with fast, parallelized data processing.
scRNA-seq has become a core technology in disease research and medicine. However, the volume of multidimensional data it produces poses challenges in analysis. Addressing this, g.nome’s end-to-end scRNA-seq solution provides pre-built workflows designed for fast, parallelized analysis of raw sequencing or counts data.

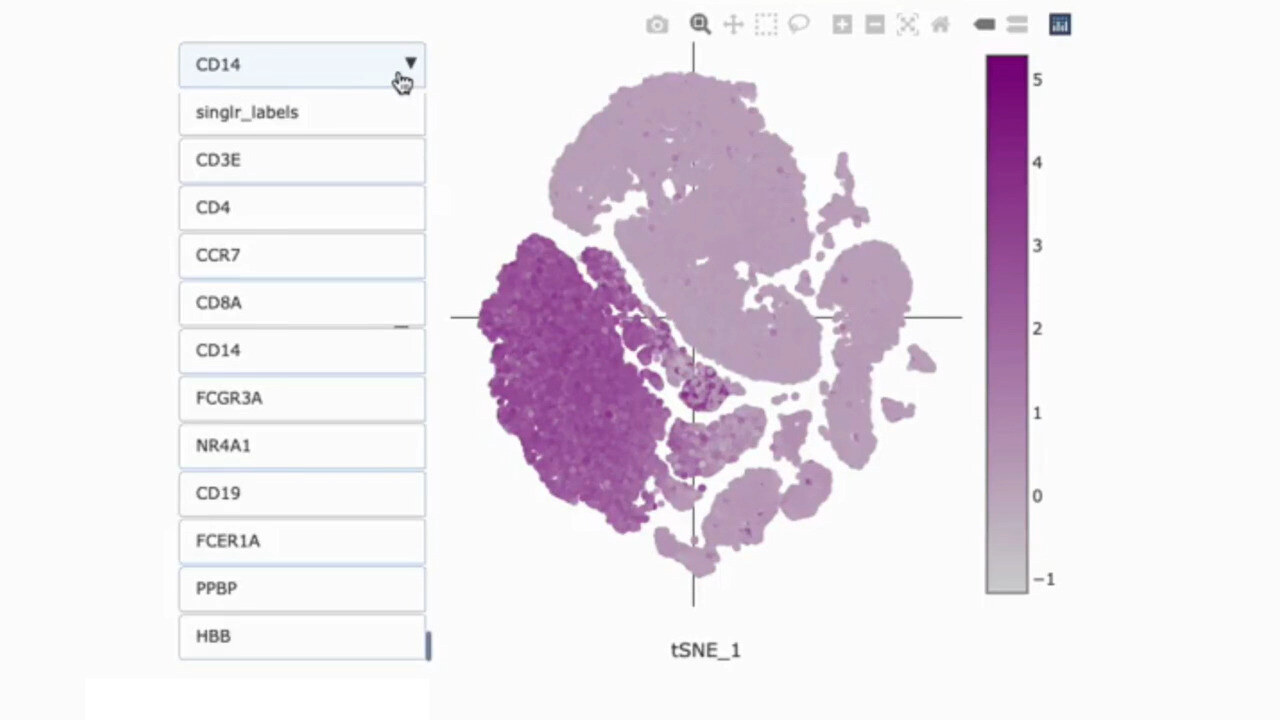
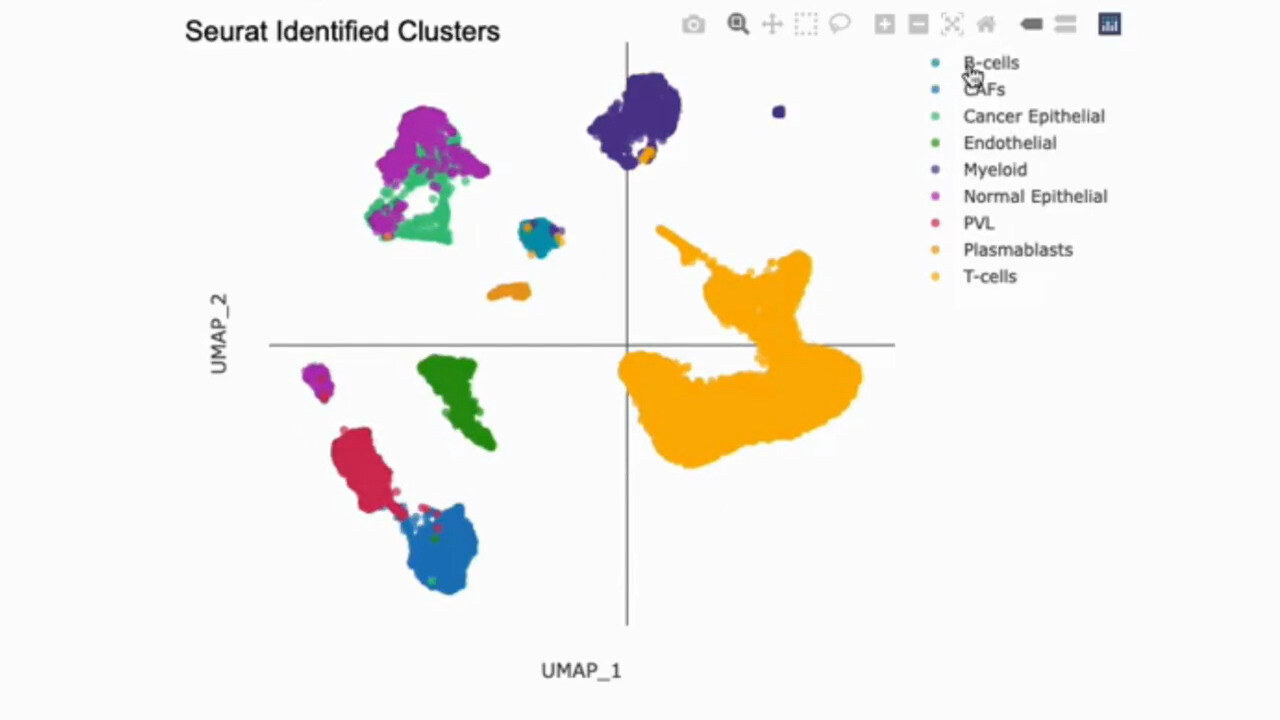
Identify Cell Types with Confidence
Cell Type Annotation
Cell type identification is critical for generating meaningful insights from single-cell RNA-seq data. g.nome utilizes advanced algorithms to accurately classify cells based on gene expression profiles.
How g.nome helps:
- Utilizes machine learning algorithms for accurate classification.
- Offers multiple reference datasets to enhance annotation accuracy.
- Provides interactive visualization tools for validation and exploration.
Unify Your Data, Eliminate Batch Effects
Batch Effect Correction and Integration
Correcting for unwanted sources of variation helps identify true biological differences. g.nome offers algorithms to correct batch effects and integrate samples for different use cases, ensuring consistency across datasets.
How g.nome helps:
- Implements state-of-the-art batch correction algorithms.
- Cloud-based platform allows integration of large datasets.
- Provides visualizations to assess the effectiveness of batch correction.
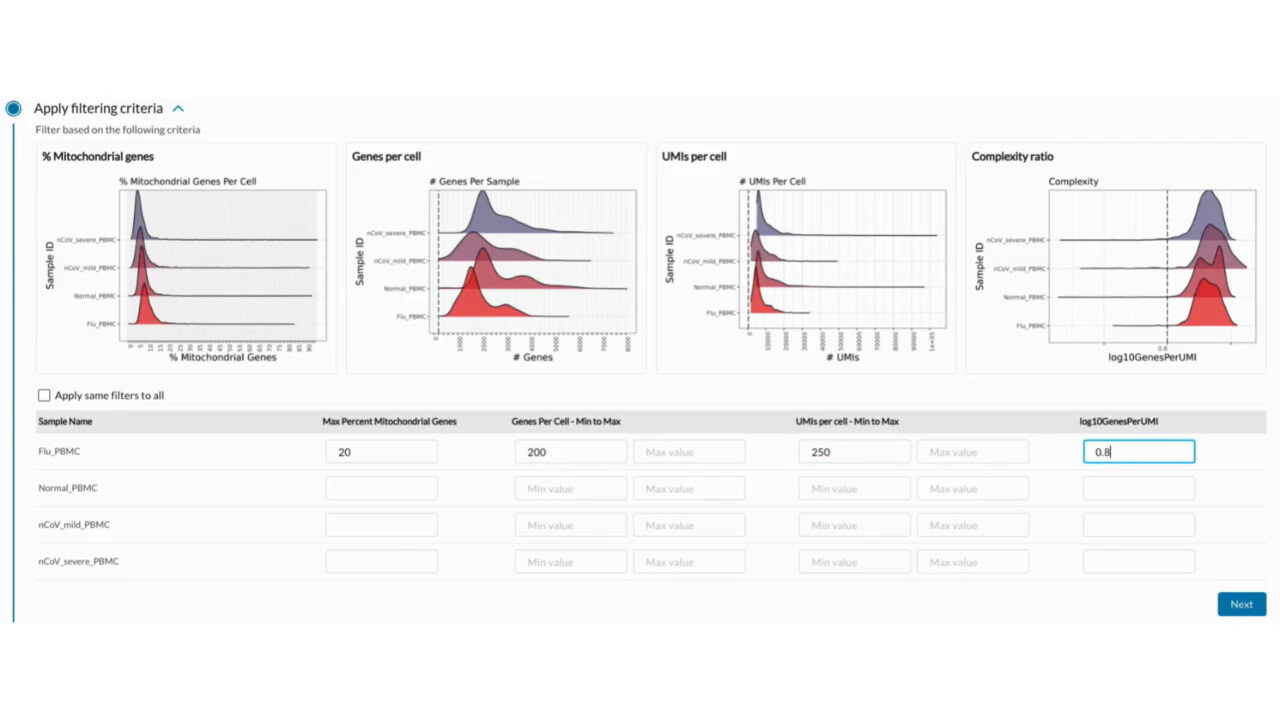
Ensure Data Integrity and Quality
Quality Control and Data Filtering
Quality control (QC) and data filtering are essential for removing noise and artifacts from single-cell RNA-seq data. g.nome automates QC processes and offers flexible filtering options to ensure high data quality.
How g.nome helps:
- Implements customizable QC metrics for assessing data quality.
- Automates data filtering based on user-defined criteria.
- Provides interactive visualizations for QC assessment and data exploration.
Identify Significant Changes with Confidence
Differential Expression Analysis
Differential expression analysis enables the identification of genes that are differentially expressed between cell populations. g.nome offers interactive visualization of gene expression across cells.
How g.nome helps:
- Implements validated tools for differential expression analysis.
- Provides intuitive visualization of significant gene expression changes.
- Generates top variably expressed genes for each cluster.
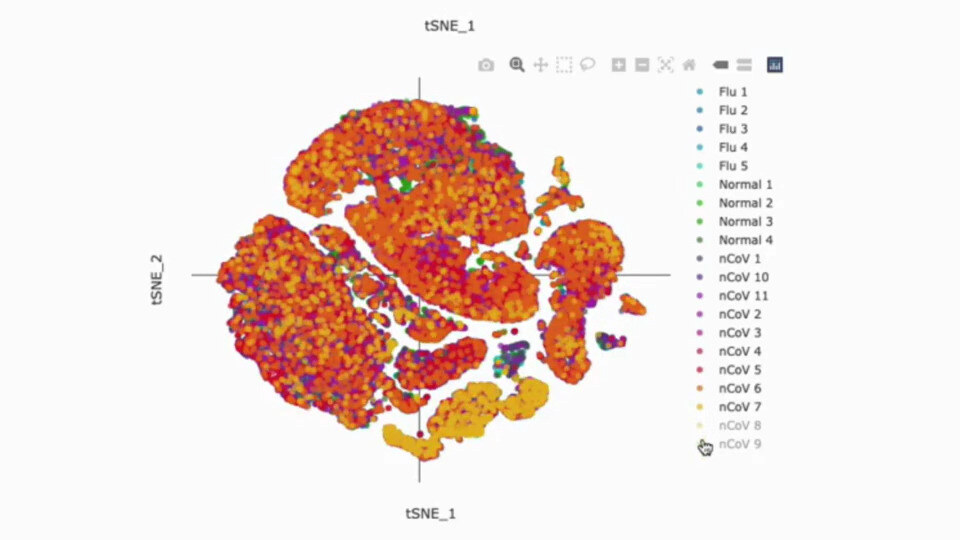
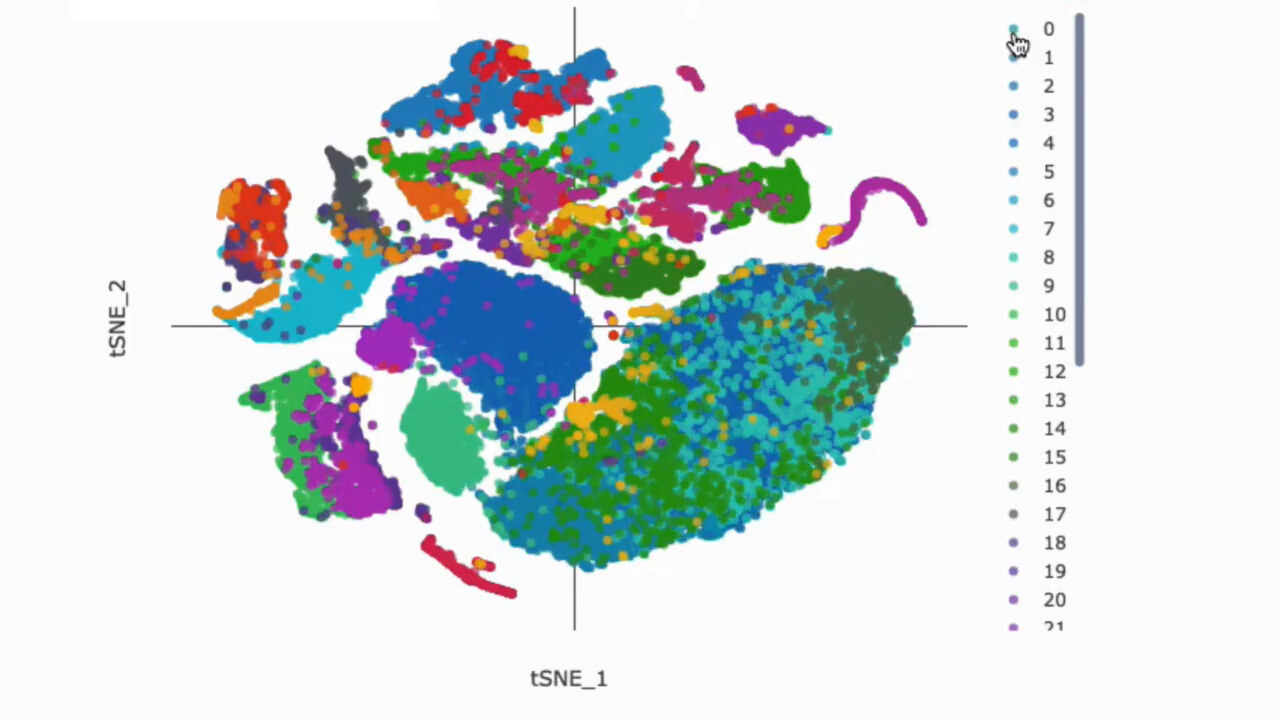
Uncover Hidden Patterns
Dimensionality Reduction and Clustering
Dimensionality reduction techniques like t-SNE and UMAP enable visualization of high-dimensional single-cell data. g.nome offers both of these techniques for efficient clustering and visualization.
How g.nome helps:
- Implements optimized algorithms for fast and accurate dimensionality reduction.
- Provides interactive visualizations for exploring clustered cell populations.
- Supports customization of clustering parameters for fine-tuning analysis.
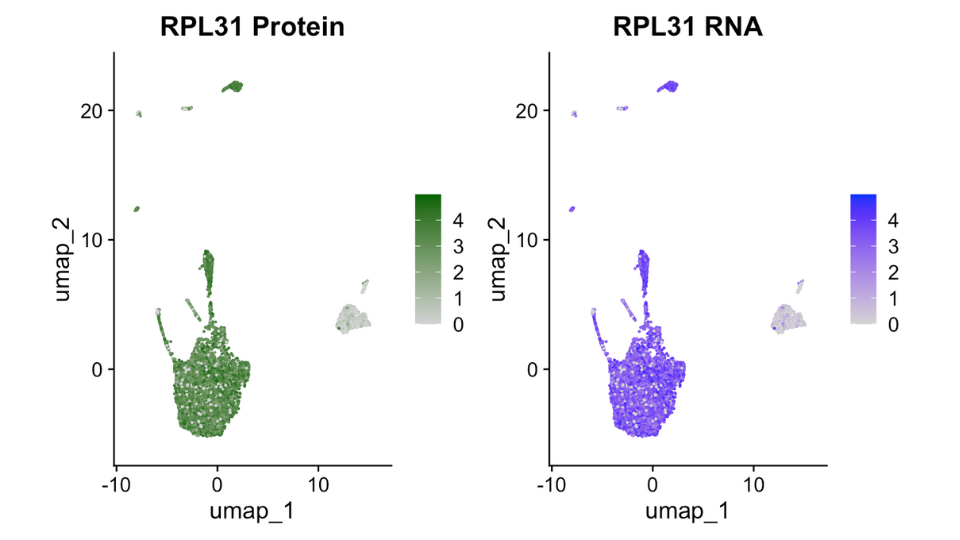
Combine Transcriptomic and Proteomic Insights
Seamless Integration of CITEseq Data
CITEseq combines single-cell RNA-seq with protein expression data, providing comprehensive insights into cell phenotypes. g.nome seamlessly integrates and analyzes CITEseq data for a holistic view of cellular states.
How g.nome helps:
- Supports simultaneous analysis of transcriptomic and proteomic data.
- Provides tools for cross-modality integration and visualization.
- Enables identification of cell subpopulations based on combined transcriptomic and proteomic profiles.
Explore the Nuclear Transcriptome
Unlock Nuclear RNA Insights with snRNA-seq data
snRNA-seq enables comparable transcriptome profiling to scRNA-seq for cells that are difficult to isolate. g.nome's single-cell platform can be used to analyze and interpret snRNA-seq data.
How g.nome helps:
- Specialized alignment settings for snRNA-seq data.
- Interactive quality control can be adjusted for nuclei.
- Fast, tunable downstream analysis.
Explore More
Bulk RNA Sequencing
Uncover gene expression changes across entire transcriptomes, enabling the identification of biomarkers and pathways associated with diseases and therapeutic responses.
Whole Genome
Delve into the entirety of the genome to identify genetic variants, structural variations, and mutations, providing comprehensive insights into genetic diseases and population genetics.

Whole Exome
Focus on the protein-coding regions of the genome to identify functional variants associated with diseases, accelerating the discovery of causative mutations and potential therapeutic targets.

Targeted Sequencing
Zoom in on specific genomic regions to accurately detect variants, structural variations, and somatic mutations, enabling precise analysis tailored to specific research or clinical questions.
BREAK THROUGH BARRIERS WITH
