Case Study: Streamlining Workflow for Quicker Builds with SelfDecode
Let’s face it: bioinformatic workflow development can be painful. Building the infrastructure, assembling the pipeline, and troubleshooting the output can take an inordinate amount of time using the conventional DIY processes. This can be risky for biotech companies trying to maintain a competitive edge because the longer it takes to build a successful workflow, the longer it takes to go to market. These bottlenecks – among other issues – are huge pain points felt large and wide in the bioinformatics community.
The Partner
SelfDecode is revolutionizing healthcare by providing personalized health recommendations – such as diet, supplement and lifestyle plans – to its customers. Using a combination of AI and machine-learning technology to conduct genomic analysis against up to 83 million genetic variants, SelfDecode is “the most accurate DNA health kit on the market”.

The Problem
Their existing genomic analysis processes were:
- Time-Consuming
Due to the time- and effort-intensive process of iteration, it took over six months for SelfDecode to develop their previous pipeline. They wanted to accelerate the development of their bioinformatic workflows, get them into production quickly, and reduce developer time needed to deploy and maintain existing workflows.
- Unreliable
Working in an expansive, fragmented environment made their pipelines difficult to reproduce and vulnerable to breaking. SelfDecode needed a solution that would ensure development of efficient, stable, and reproducible bioinformatic workflows, scalable runs, and ease of maintenance.

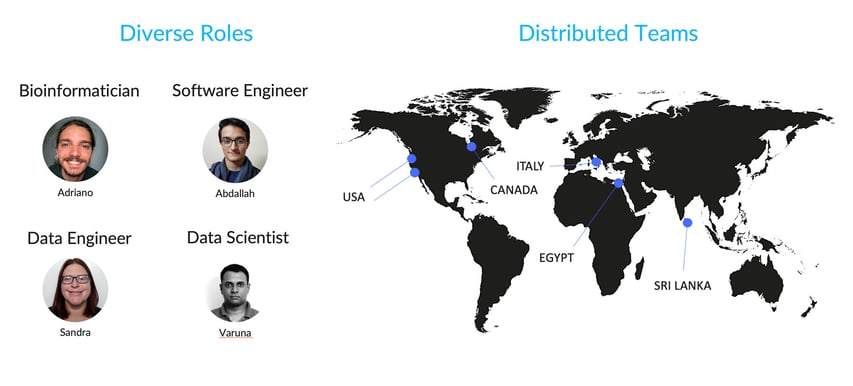
- Counter-Collaborative
Despite having a diversely talented team spread across four continents, SelfDecode had to rely solely on their bioinformatician for the heavy lifting of pipeline development. In order to facilitate better collaboration, they needed a common language to be able to involve users with varying levels of bioinformatics expertise and capabilities in the workflow development process.
The Solution
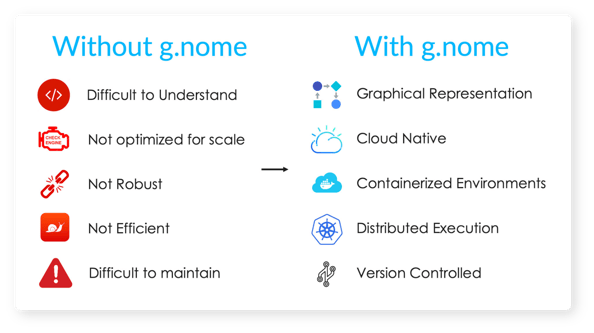
Almaden Genomics introduced SelfDecode to g.nome®, a cloud-native platform designed to streamline genomic workflows.
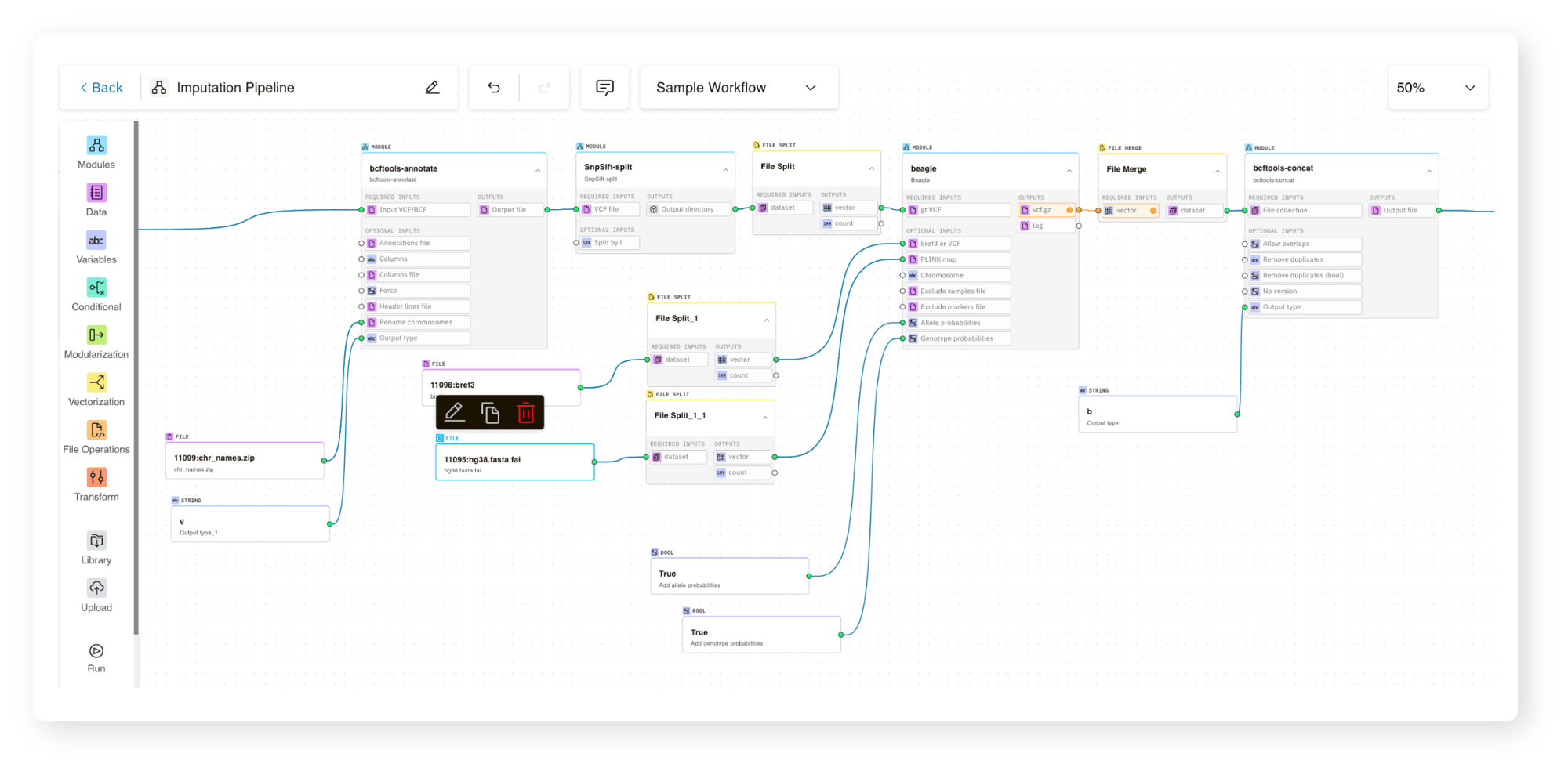
Fortunately, none of the coding and assembly that their team had previously done would go to waste. With g.nome, SelfDecode was able to:
- Import their own data (cloud and local)
- Bring on existing tools
- Add their unique custom code
They then continued to build their pipeline leveraging g.nome’s powerful suite of features, including vectorization and conditional execution.
The Results
Using g.nome’s powerfully simple workflow builder, SelfDecode reduced the pipeline build time from 13 weeks to 2 hours. g.nome not only enabled their users to spot problems easier and iterate faster using the graphical pipeline builder, but the visual representation of workflows also facilitated easy collaboration with non-bioinformaticians on the team.
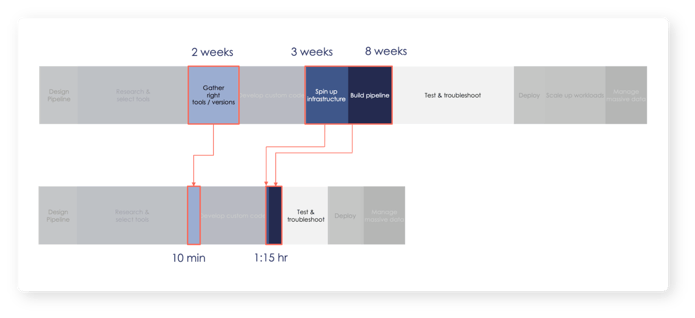

- Avoiding the need for complex setup by their IT team with the use of g.nome’s optimized cloud, SelfDecode saw overall computational efficiency with 3X faster runtime at a fraction of the cost.

- The reduction in build and run time led to an estimated direct savings of $20K in FTE costs.

- Real-time collaboration among SelfDecode’s multi-disciplinary team was made easier with g.nome, even across five different time zones and four continents.
- g.nome provided a single platform to transition prototypes from R&D to production as reusable, scalable, assets.
Conclusion
SelfDecode found that by using g.nome, they could speed up their development time while also saving costs. In addition, g.nome represents an opportunity for their entire team – not just bioinformaticians and software engineers – to actively collaborate on new workflows. They see a long-term value in g.nome and look forward to using it as part of their key development process.
Don’t take our word for it. Here’s what the SelfDecode team had to say:


