TARGETED SEQUENCING
Achieve unparalleled precision in genetic variant analysis.
Our platform provides a comprehensive suite of tools and algorithms for precise detection and analysis of genetic variants within specific regions of interest. Whether you are focusing on variant calling, CNV/Indel analysis, somatic sequencing, or germline sequencing, g.nome offers the accuracy, customization, and visualization you need to make groundbreaking discoveries.
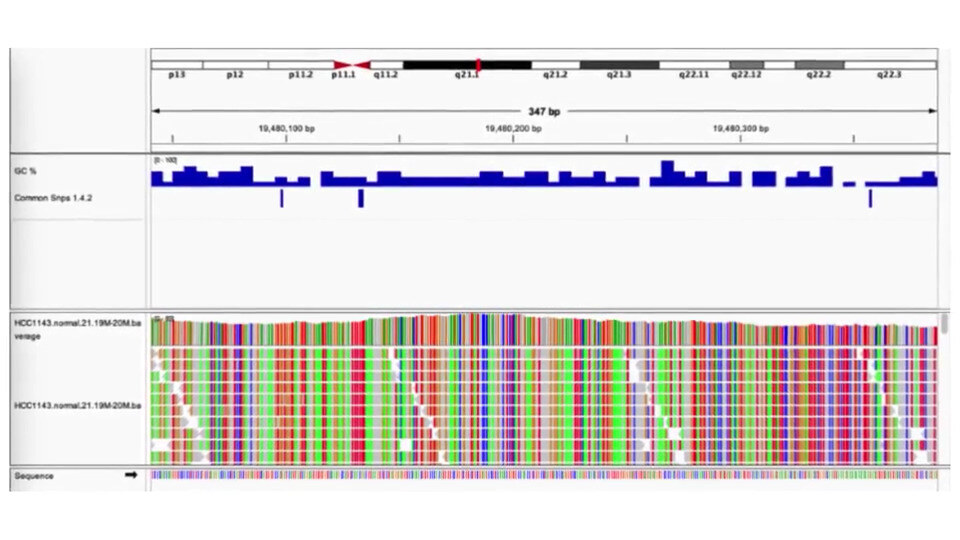
Unravel Somatic Mutations with Confidence
Somatic Sequencing Analysis
Somatic sequencing in targeted sequencing identifies genetic mutations specific to tumor cells or other somatic tissues within the regions of interest. g.nome offers specialized pipelines and analysis tools for accurate somatic variant detection and interpretation from targeted sequencing data.
How g.nome helps:
- Implements robust workflows for somatic variant calling and filtering.
- Integrates with cancer databases and annotation tools for variant interpretation.
- Provides customizable reporting features for communicating findings in research or clinical settings.
.jpg?width=960&height=540&name=2024%20New%20Website%20GIFs(4).jpg)
Explore Inherited Genetic Variants with Ease
Germline Sequencing Analysis
Germline sequencing in targeted sequencing identifies genetic variants inherited from parents within the regions of interest. g.nome offers specialized pipelines and analysis tools for accurate germline variant detection and interpretation from targeted sequencing data.
How g.nome helps:
- Implements robust algorithms for germline variant calling and annotation.
- Provides integration with population databases to interpret the frequency and clinical significance of germline variants.
- Offers customizable reporting features for communicating findings in research or clinical settings.

Identify Genetic Variants with Precision
Variant Calling
Variant calling in targeted sequencing focuses on detecting genetic variations within specific regions of interest. g.nome employs advanced algorithms to accurately call variants, including single nucleotide variants (SNVs) and small insertions/deletions (Indels), from targeted sequencing data.
How g.nome helps:
- Utilizes specialized variant calling algorithms optimized for targeted sequencing data.
- Provides customizable filtering options to prioritize variants of interest.
- Offers interactive visualization tools for variant exploration and validation.
Detect Structural Variations with Ease
CNV and Indel Analysis
CNV and Indel analysis in targeted sequencing identifies structural variations within the regions of interest. g.nome offers specialized tools to accurately detect and analyze CNVs and Indels from targeted sequencing data, providing insights into genomic alterations.
How g.nome helps:
- Implements tailored algorithms for CNV and Indel detection within targeted regions.
- Provides customizable visualization tools for exploring CNV and Indel profiles.
- Integrates annotation databases to interpret the functional impact of detected variants.
Explore More
Single-Cell RNA-seq
Explore the complexity of cellular heterogeneity and gene expression patterns at single-cell resolution, enabling deeper insights into cell types and states in diverse biological systems.
Bulk RNA Sequencing
Uncover gene expression changes across entire transcriptomes, enabling the identification of biomarkers and pathways associated with diseases and therapeutic responses.
Whole Genome
Delve into the entirety of the genome to identify genetic variants, structural variations, and mutations, providing comprehensive insights into genetic diseases and population genetics.

Whole Exome
Focus on the protein-coding regions of the genome to identify functional variants associated with diseases, accelerating the discovery of causative mutations and potential therapeutic targets.
BREAK THROUGH BARRIERS WITH
