BULK RNA-SEQ
Enhance your bulk RNA-Seq research with advanced analysis solutions.
g.nome provides comprehensive tools for bulk RNA-Seq analysis, including differential expression, pathway analysis, gene set enrichment, gene fusion detection, alternative splicing analysis, and quality control. Our platform integrates robust algorithms, customizable workflows, and interactive visualizations to uncover meaningful insights from your data, ensuring accurate and reliable results throughout your research journey.
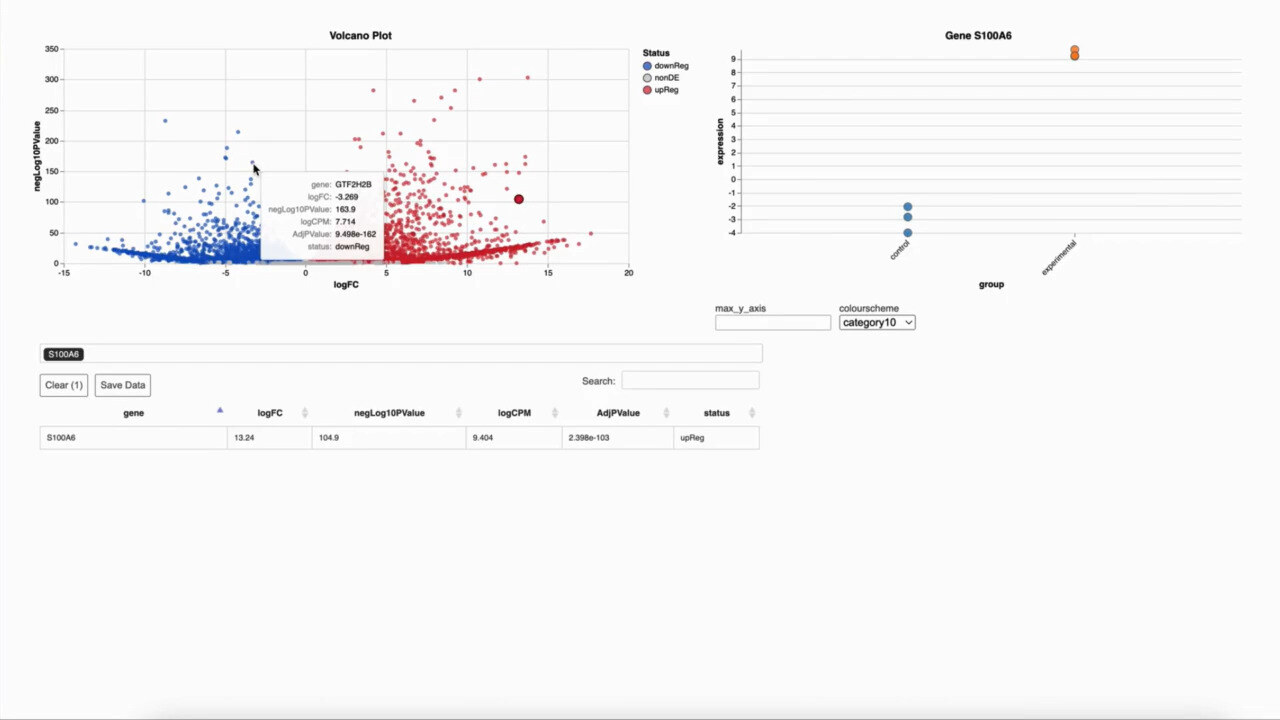
Identify Significant Gene Expression Changes
Differential Expression Analysis
How g.nome helps:
- Implements statistical methods for accurate differential expression analysis.
- Provides customizable options for controlling false discovery rates.
- Offers interactive visualizations for exploring expression changes across conditions.
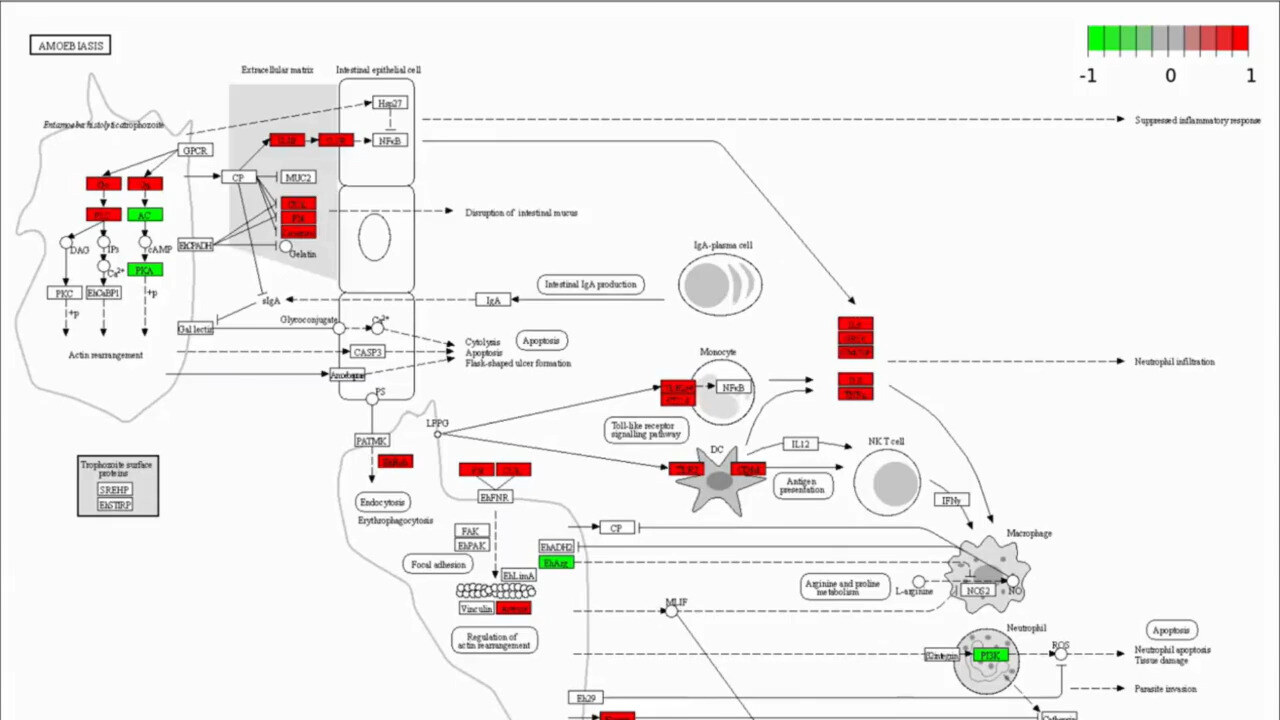
Discover Functional Significance
Pathway Analysis
Pathway analysis identifies biological pathways enriched with differentially expressed genes, providing insights into underlying biological processes. g.nome integrates pathway databases and offers intuitive tools for pathway analysis.
How g.nome helps:
- Integrates popular pathway databases for comprehensive analysis.
- Provides visualization of enriched pathways and associated genes.
- Offers statistical assessment of pathway enrichment significance.
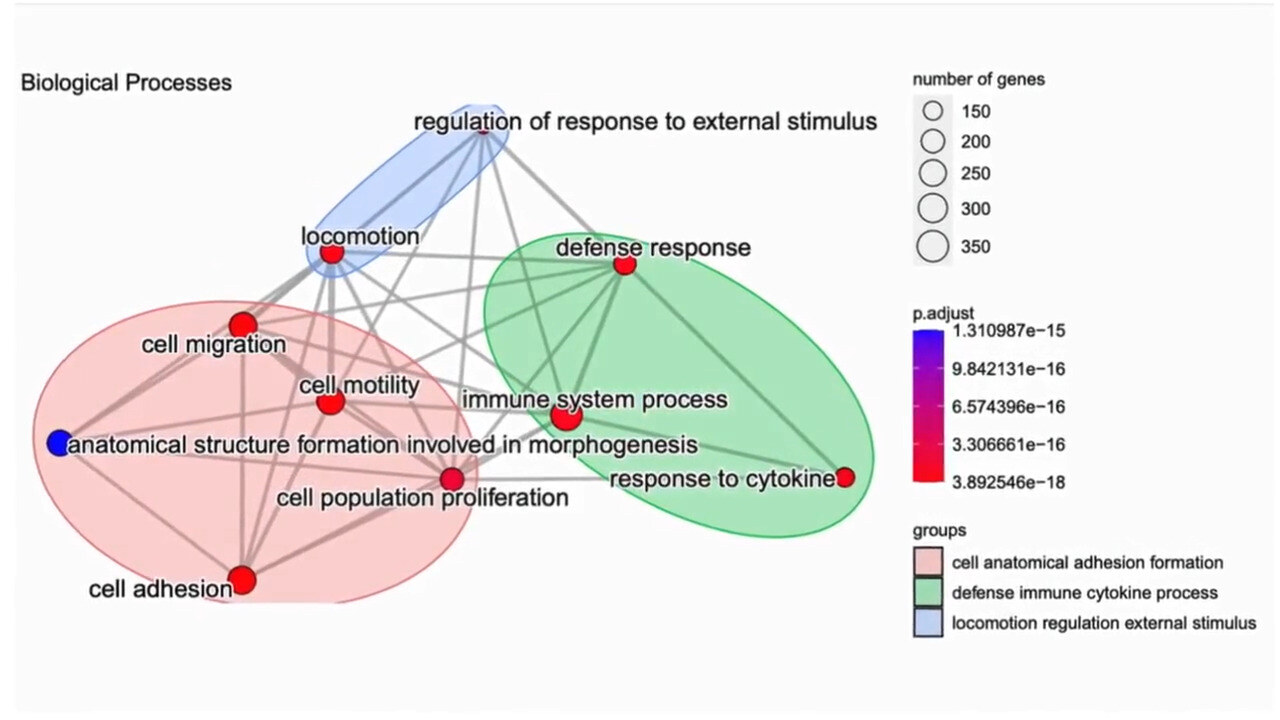
Identify Functional Signatures in Your Data
Gene Set Enrichment Analysis
How g.nome helps:
- Implements a variety of statistical methods for gene set enrichment analysis.
- Provides curated gene set databases for different biological contexts.
- Offers visualization tools to interpret enrichment results in a biological context.
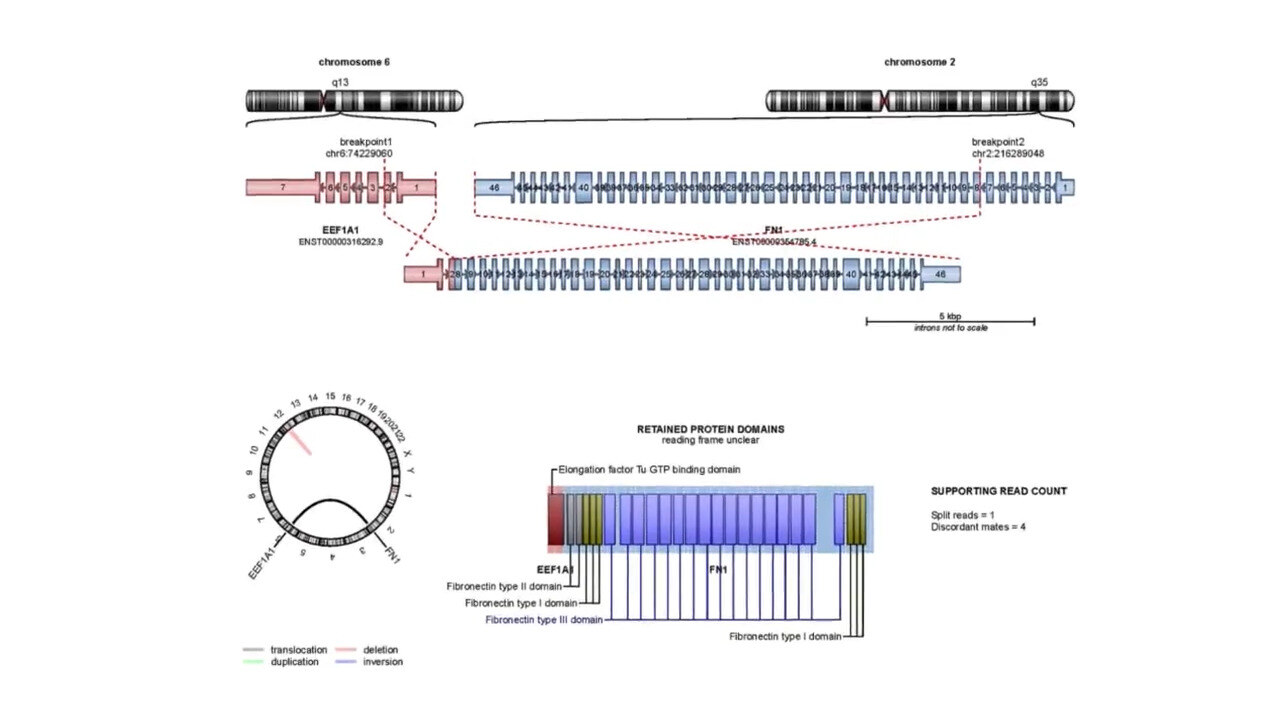
Detect Structural Variations Impacting Gene Expression
Gene Fusion Analysis
How g.nome helps:
- Utilizes advanced algorithms to accurately detect gene fusion events.
- Provides annotations and visualizations to characterize fusion events.
- Offers integration with genomic databases to identify known and novel fusion events.
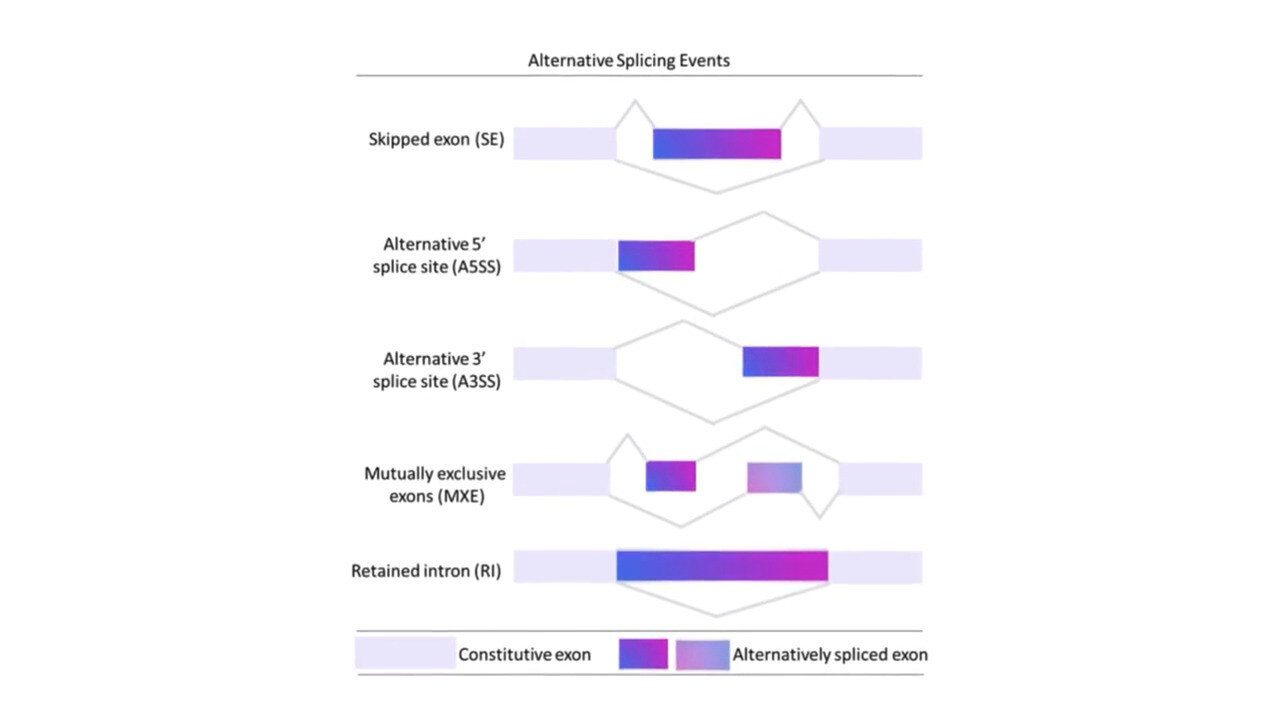
Explore Transcript Diversity in Depth
Alternative Splicing
Alternative splicing generates multiple mRNA isoforms from a single gene, contributing to transcriptome diversity. g.nome offers tools to identify and analyze alternative splicing events, providing insights into gene regulation and function.
How g.nome helps:
- Implements algorithms to detect and quantify alternative splicing events.
- Provides visualization tools to explore splicing patterns across samples.
- Offers annotation databases to characterize splicing events and their functional impact.
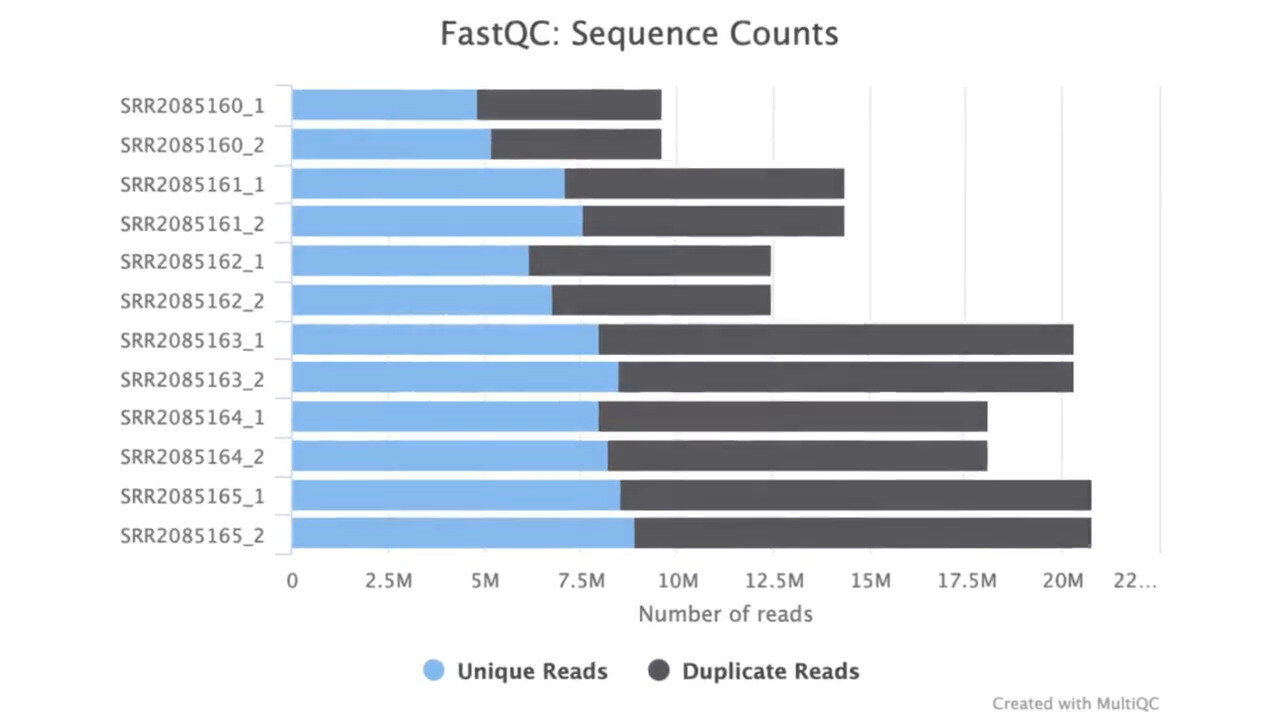
Ensure Data Integrity Every Step of the Way
Efficient QC for Reliable Results
Quality control (QC) is essential for ensuring the reliability and validity of bulk RNAseq data. g.nome automates QC processes and provides comprehensive QC metrics to assess data quality throughout the analysis pipeline.
How g.nome helps:
- Implements customizable QC metrics to assess data quality.
- Automates detection and removal of low-quality samples or outliers.
- Provides interactive visualizations for QC assessment and data exploration.
Explore More

Single-Cell RNA-seq
Explore the complexity of cellular heterogeneity and gene expression patterns at single-cell resolution, enabling deeper insights into cell types and states in diverse biological systems.

Whole Genome
Delve into the entirety of the genome to identify genetic variants, structural variations, and mutations, providing comprehensive insights into genetic diseases and population genetics.

Whole Exome
Focus on the protein-coding regions of the genome to identify functional variants associated with diseases, accelerating the discovery of causative mutations and potential therapeutic targets.

Targeted Sequencing
Zoom in on specific genomic regions to accurately detect variants, structural variations, and somatic mutations, enabling precise analysis tailored to specific research or clinical questions.
BREAK THROUGH BARRIERS WITH
