Differential Expression in Colorectal Cancer via Bulk RNA seq
Colorectal cancer (CRC) is a major global health concern1. Personalized medicine in CRC treatment is gaining significance, allowing tailored therapies based on individual genetic, molecular, and environmental factors. These advances, particularly in RNA sequencing and genomics, enable a precise understanding of CRC at the molecular level. This knowledge aids in identifying biomarkers and therapeutic targets, ushering in more effective, patient-specific treatments. Personalized medicine in CRC signifies a shift towards precise, data-driven healthcare strategies.
CRC is a complex disease influenced by genetic and epigenetic factors, leading to therapy heterogeneity and drug resistance. Personalized medicine in CRC categorizes patients based on their unique genetic and epigenetic characteristics, optimizing therapeutic approaches2. This includes chemotherapy and targeted therapy, focusing on specific biomarkers and genetic profiles. Targeted drugs like Cetuximab and chemotherapy agents like Fluoropyrimidines and Oxaliplatin exemplify personalized CRC treatments, marking a significant advancement in treatment strategies.
The study titled "NFKB2 mediates colorectal cancer cell immune escape and metastasis in a STAT2/PD-L1-dependent manner" explores NFKB2's role in CRC. It reveals that NFKB2 upregulation induces CD8+ T cell exhaustion and increases PD-L1 expression, potentially suppressing immune surveillance3. Targeting NFKB2 could enhance immunotherapies' effectiveness, highlighting personalized medicine's significance in CRC treatment. The study underscores the potential of RNAseq data in developing targeted therapies.NFKB2's role in CRC, particularly in immune escape and metastasis, is crucial. Upregulated NFKB2 can lead to immune suppression by affecting CD8+ T cells and PD-L1 expression. Targeting NFKB2 is vital in CRC treatment, potentially improving immunotherapies.
Analyzing RNAseq data can provide nuanced insights into CRC's molecular mechanisms, identifying new therapeutic targets, enhancing tumor-immune interactions understanding, and developing personalized treatments. Precision medicine in oncology leverages molecular data for tailored therapies, exemplifying its growing importance.
OBJECTIVES
- Investigate NFKB2's Impact: Explore NFKB2's role in CRC immune escape and metastasis.
- Utilize Advanced Bioinformatics: Employ the g.nome® platform for in-depth genetic and epigenetic analysis.
- Quality Control and Data Integrity: Ensure high-quality data through stringent QC measures.
- Molecular Analysis: Conduct differential expression and pathway analyses to identify CRC biomarkers and therapeutic targets.
- Visualize Gene Interactions: Use chord plot visualizations to interpret complex gene relationships and their implications for CRC progression and treatment.
METHODOLOGY
g.nome's intuitive bioinformatics platform simplifies the process of importing and analyzing biological data with its drag-and-drop tooling. Starting with the retrieval of an SRA dataset, users can easily drag the desired SRA accession list into the workspace to initiate the sra-toolkit-prefetch. This module seamlessly downloads the dataset without any complex command-line instructions.
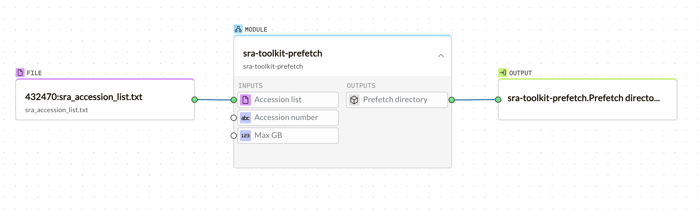
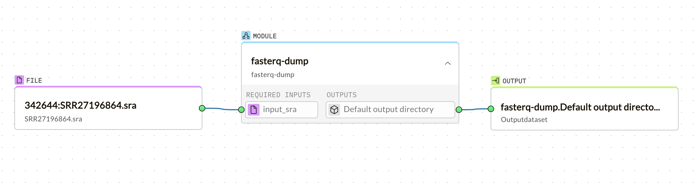
Following the download, the fasterq-dump module is just a drag away, converting SRA files into FASTQ format, ready for analysis.
This user-friendly approach is extended to the RNAseq pipeline, where the workflow is laid out in a straightforward manner. Users can follow the visual cues to conduct quality control checks, alignments, and differential expression analysis.
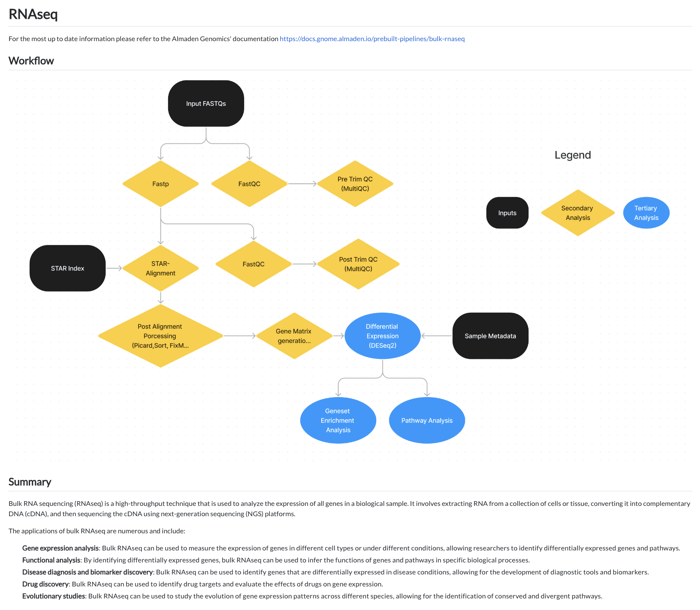
For further guidance, a video file provides a step-by-step walkthrough, ensuring that even those with minimal bioinformatics experience can navigate the platform with ease and conduct comprehensive RNAseq analyses. This efficient system effectively reduces the barrier to entry for conducting complex genomic research.
RESULTS
Not only is it easy to set up a run in g.nome, but it also runs efficiently in a cloud-elastic fashion. From defining the inputs to examining the results can take less than an hour. See below for key usage statistics for this analysis on g.nome:
- Analysis Time: 54 minutes, 46 seconds
- Qty of Data Processed: 12.5GB SRA data, 20.42GB FASTQ data
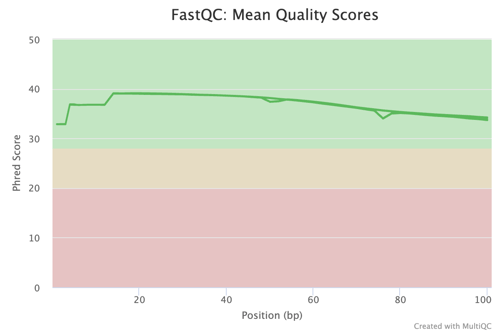
Quality Control (QC)
In any high-stakes research such as colorectal cancer studies, the integrity of data is crucial. The g.nome platform ensures the utilization of high-quality data by implementing rigorous QC protocols. This commitment to data quality is evidenced by the FastQC Mean Quality Scores figure, which indicates consistent high-quality scores across the sequence reads.
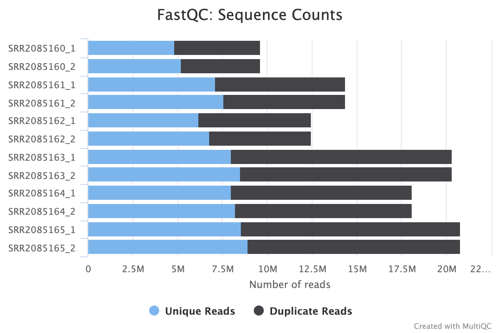
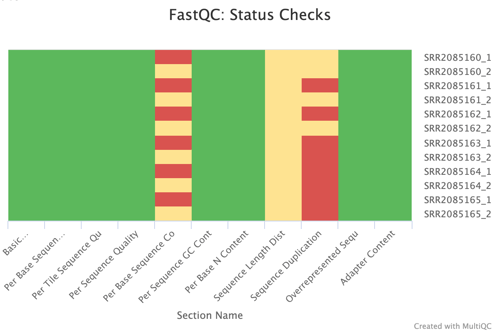
Sequence Counts and Status Checks
The FastQC Sequence Counts chart showcases the number of unique and duplicate reads, providing a quantitative measure of the sequencing depth and richness of the dataset. The Status Checks graph offers a color-coded overview of various QC metrics, allowing for quick identification of potential issues that may require attention.
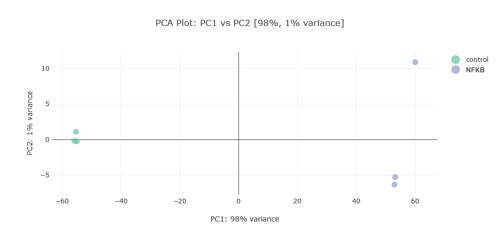
Alignment and Principal Component Analysis (PCA)
The alignment tools within g.nome are pivotal for highlighting genetic discrepancies between healthy and cancerous tissue, a crucial step for downstream analyses. Furthermore, the PCA plot illustrates the variance within the sample set, underlining the robustness of the dataset and the ability to discriminate between different genetic expressions which may be pivotal in understanding colorectal cancer.
The integration of these tools into the g.nome platform not only ensures data integrity but also enhances the efficiency of the research workflow, from initial quality assessment to in-depth genetic analysis. The g.nome platform is a pivotal resource in the quest to unravel the genetic intricacies of colorectal cancer and advance the development of targeted therapies.
Differential Expression (DE)
Our DE analysis is pivotal in order to understand colorectal cancer at the molecular level. Utilizing the sophisticated visualization capabilities of the g.nome platform, we delve into the complexities of gene expression differences. Heatmaps offer us a color-coded representation of gene expression levels across different samples, highlighting genes that are significantly up or downregulated — a feature clearly demonstrated in the clustering of genes and samples. This visualization is particularly telling, as it brings to light the top 25 differentially expressed genes in a coherent and interpretable format.
The volcano plot further complements these findings by contrasting the magnitude of expression changes (log fold change) against the statistical significance (negative log of the p-value), thereby pinpointing genes of interest with dramatic changes in expression levels. Such genes emerge as potential biomarkers or therapeutic targets. In the context of our colorectal cancer dataset, identifying genes like ANXA1 and KRT5 as significantly upregulated, which is in line with their known involvement in cancer progression and metastasis. AR and STAT6 also emerge from the data, prompting a deeper investigation into their specific roles in CRC.
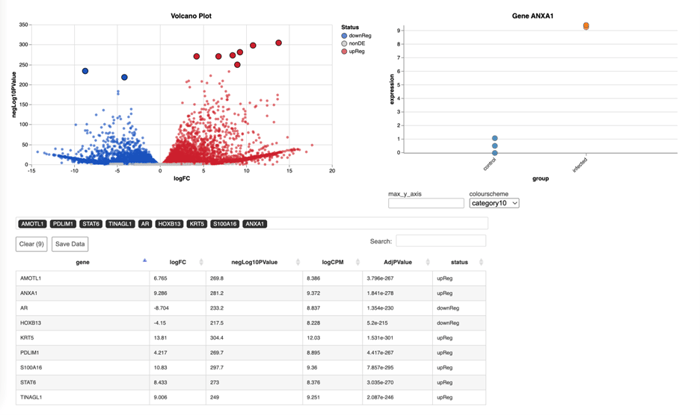
.png?width=650&height=321&name=newplot(2).png)
Conclusion
The strength of g.nome lies in its capacity to rapidly integrate data and execute complex bioinformatics protocols with ease, enabling researchers to focus on interpreting results rather than managing data. Visual tools such as heatmaps and volcano plots distill complex data sets into clear, concise visual representations, highlighting pivotal genes implicated in colorectal cancer.
These visual aids are particularly adept at revealing genetic expressions and mutations relevant to colorectal cancer pathogenesis, offering scientists the ability to swiftly identify potential targets for further study. As a result, g.nome stands out as a valuable asset in the field of genetic research, fostering an environment where significant biological discoveries are made more accessible and less time-intensive.
References
- Rahbari NN et al. "Personalized Medicine in Colorectal Cancer: A Review of Current Status and Future Perspectives." PubMed, 2020. DOI: 10.2174/1389450119666180803122744. PubMed.
- Tieng FYF et al. "Deciphering colorectal cancer immune microenvironment transcriptional landscape on single cell resolution - A role for immunotherapy." Front Immunol. 2022 Aug 10;13:959705. doi: 10.3389/fimmu.2022.959705. PMID: 36032085; PMCID: PMC9399368. Frontiers in Immunology.
- Xu L et al. "NFKB2 Mediates Colorectal Cancer Cell Immune Escape and Metastasis in a STAT2/PD-L1-Dependent Manner." NCBI GEO, NCBI GEO.